The U.S. healthcare system was not prepared for a health crisis of the magnitude of the COVID-19 pandemic. Hospitals are working to facilitate widespread distribution of information within their organization and to local, state, and federal authorities to successfully manage this novel infection. EHRs and Lab Information Systems (LISs) have become public health tools for disease surveillance and management.
Due to signification variation in EHR data, informatics tools are needed to define patients with suspected SARS-Cov2 Infection and confirmed COVID-19 infection. With the aim of building an extensible model for a COVID-19 database, Health Catalyst has built a detailed approach that leverages a heuristic methodology for capturing both confirmed and suspected cases.
Health Catalyst has proposed value sets that define two patient cohorts for the registry for confirmed and suspected COVID-19 patients, stratified further into three levels of confidence: high confidence suspected, moderate confidence suspected, and low confidence suspected.
 Download
Download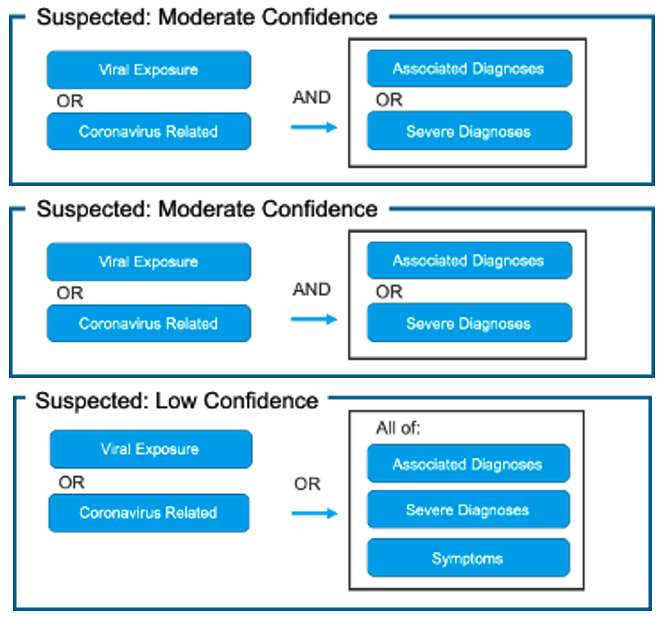

Since the World Health Organization (WHO) declared the coronavirus disease 2019 (COVID-19) as a pandemic on March 11, 2020, the United States has become the epicenter of the disease.1 While the numbers continue to increase, there are 4,710,282 confirmed cases1, surpassing the national case numbers of Spain, Italy, Germany, France, and China. This pandemic has highlighted the fact that the U.S. healthcare system is not prepared for a health crisis of this magnitude.2 EHRs and Lab Information Systems (LISs) have become public health tools for disease surveillance and management. Hospitals are working to facilitate widespread distribution of information within their organization and to local, state, and federal authorities to successfully manage this novel infection.
Additionally, reports of sparse testing kits, ventilators, and personal protective equipment (PPE) drive an urgent need for a means to calculate surge capacity requirements.3 EHR data has significant variation and informatics tools are needed to define patients with suspected SARS-Cov2 Infection and confirmed COVID-19 infection. With the aim of building an extensible model for a COVID-19 database, we have built a detailed approach that leverages a heuristic methodology for capturing both confirmed and suspected cases.
Based on the guidance provided by the CDC,4,5 WHO,6, 7 AMA,8 NLM VSAC,9 AAPC10 and published literature, we propose value sets that define two patient cohorts for the registry of confirmed and suspected COVID-19 patients, stratified further into three levels of confidence: high confidence suspected, moderate confidence suspected, and low confidence suspected. These value sets are based on ICD-10-CM codes (Supplemental Table 2) and will continue to evolve as we understand more about COVID-19, and as clinical practices change over time. Using the four patient cohorts for the registry defined using the value sets above, patient care outcomes can be studied.
We developed modified indicators that classify diagnosis codes into one of six value-sets (Diagnosis-Grain) and added additional logic on top of this to classify a patient into one of four diagnosis categories (Patient-Grain) intended to represent varying levels of diagnostic certainty. This was developed using Health Catalyst Touchstone® data with the intent to capture suspected C-19 patients not readily identifiable by lab tests and preceding widespread adoption of the ICD-10 code U07.1 for confirmed COVID-19, released for use on April 1, 2020.
Diagnosis-Grain:
1) Confirmed COVID-19
2) Viral exposure
3) Coronavirus-related
4) Associated COVID-19 Diagnoses
5) Severe Associated COVID-19 Diagnoses
6) COVID-19 Symptoms
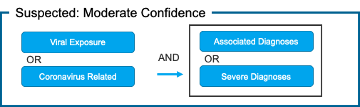
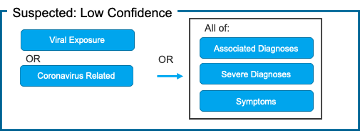
Patient-Grain: [Figure 1]
1) Confirmed COVID-19
2) Suspected: High Confidence
3) Suspected: Moderate Confidence
4) Suspected: Low Confidence
Fig. 1. Proposed Use of Value Sets for Generating COVID-19 Patient Cohorts Using COVID-19 Categorization Logic

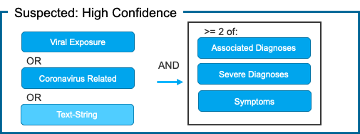
The confirmed cohort removes ICD-10 Code B97.29 “Other coronavirus as the cause of diseases classified elsewhere” as this code captures all coronaviruses (including and in addition to COVID-19). People around the world are commonly infected with human coronaviruses 229E, NL63, OC43, and HKU1. There are additional human coronaviruses that are captured through the use of ICD-10 code B97.2. In our diagnosis logic, patients with this code are classified as “Suspected: High Confidence” if they do not have a positive non-COVID-19 coronavirus lab test.
Associated adds additional ICD-10 codes capturing the same or similar diagnoses as those documented in the CDC Guidelines, but extends beyond the specific ICD-10 codes documented in CDC Guidance. This is to account for varying usage of ICD-10 codes by clinicians across health systems. This value-set is intended to capture diagnoses characteristic of COVID-19 (moderate in severity, as mild symptoms and severe complications are captured in separate value-sets). Two codes have been moved to the Health Catalyst Severe Diagnosis value-set for purposes of use in the patient-grain diagnosis logic.
Health Catalyst value-sets place the below codes in different value-sets for the purposes of allowing the patient-grain diagnosis logic to require additional elements to increase confidence that these codes are not representing non-C-19 coronaviruses. Clinically, these codes are intended to be used in conjunction with other diagnosis codes. Our value-sets do not include ICD-10 code Z03.818 “Encounter for observation for suspected exposure to other biological agents ruled out” as the guidance for this code is to be used to document cases ruled out.
People around the world are commonly infected with human coronaviruses 229E, NL63, OC43, and HKU1. Please see https://www.cdc.gov/coronavirus/types.html for additional human coronaviruses that are captured through use of ICD-10 codes B34.2 and B97.2.
The codes in this value-set are found in the CDC Value-set ‘Suspected’ but are moved to their own value-sets for the purposes of the Patient-Grain logic.
This value-set is intended to be used in conjunction with other value-sets in the Patient-Grain Diagnosis Logic to help identify those patients who have developed severe symptoms/complications associated with C-19 likely requiring hospital-level care, in addition to other known C-19 symptoms and/or associated diagnoses. This is a compilation of some of the most commonly noted severe complications of COVID-19 noted in the literature.
This value-set is intended to be used in conjunction with other value-sets in the Patient-Grain Diagnosis Logic to help identify those patients who have symptoms of C-19 in addition to other known C-19 severe complications and/or associated diagnoses. This is a compilation of some of the most commonly noted symptoms and emerging symptoms of COVID-19 noted in the literature.
Due in part to the critical diagnostic importance of lab testing, lab test result data are key to understanding the clinical state of patients as well as surveillance of patient populations. However, in EHR systems lab test result data are often stored with local codes or strings for lab test types and test result values rather than codes from widely standardized terminologies such as LOINC (Logical Observation Identifiers Names and Codes).19 In such cases, there is a need to ascertain the types of the lab tests and the meanings of the result values automatically over a large volume of lab result data without the benefit of a uniform standard lab terminology across the various EHR systems from which the lab result data are sourced.
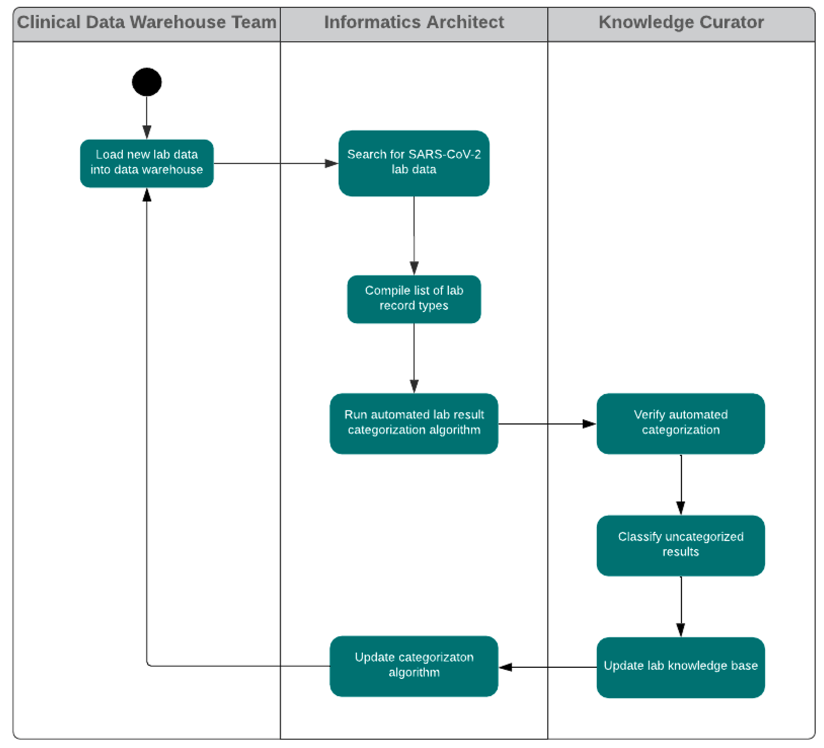
In this work, a lab test knowledge curation workflow (fig. 2) was established in order to provide a knowledge base for recognizing lab test types and understanding lab result values as expressed in the lab result data records from multiple EHR systems. This lab test knowledge curation workflow is shown in the following diagram. This knowledge base, accumulated from many lab result records across multiple EHR systems, allows for automated categorization of lab results (positive, negative, pending, ambiguous, test problem, unmapped) and lab test types (detection of SARS-CoV-2 material, detection of antibody to SARS-CoV-2). This classification system is of great convenience for use in analytics to find patterns of interest in, for example, the data of patients who are confirmed or suspected COVID-19 cases. The harmonized set of lab result values allows for rapid automated use of readily comprehensible data (i.e., positive and negative result values), identification of results that may be usable with additional data curation (i.e., unmapped), and the ability to select for patients whose tests may still be in process or unavailable (i.e., pending).
Would you like to learn more about this topic? Here are some articles we suggest: